Introduction
Medical Imaging AI solutions need to be deployed at scale, sometimes on new hardware platforms, but also on legacy platforms. This is feasible if inference can be performed efficiently and easily on IA. Once the DNN model is optimized using Intel OpenVINO , it can be deployed on numerous Intel Architecture using the same API. This provides developers the flexibility to choose the right architecture as per their performance, price, and power requirements.
We showcase the benefits of Intel® Distribution of OpenVINO for Medical Imaging AI through the Brain Tumor Segmentation (BTS) reference implementation, that uses of Intel® Distribution of OpenVINO™ Toolkit to detect and segment brain tumors in MRI images. This implementation can perform segmentation on CPU (Core and Xeon), iGPU, and VPU in an efficient manner with no code change when switching IA platforms.
Given OpenVINO interoperability and the diversity of IA hardware solutions from which developers can choose from, Intel provides platforms like Intel(R) DevCloud for Edge to developers as playground to build, test, and optimize their solutions. It provides a free cluster of Intel processors (CPU, iGPU, VPU) to test their Medical Imaging model and choose the best hardware as per their workloads. The BTS sample application is available on the DevCloud for the Edge for developers to use as it is or use it as a base-code to bootstrap their customized solution. Intel also provides Intel(R) Developer Catalog which offers reference implementations and software packages to build modular applications using containerized building blocks. Using the containerized building blocks the developers can rapidly develop deployable solutions for the edge. We have a BTS reference implantation in the Intel Developer catalog for developers to get a container-based solution to use as is or as base code for their implementations.
The Challenge
Developers are looking for options to deploy Medical Imaging AI workloads at scale. Developers are generally constrained by high cost of GPU, lack of sample code/solutions to use as a base for their application to not start from scratch or reinvent the wheel so to say. Developers seem to be bound by the performance that the popular framework like TensorFlow or PyTorch can offer them. Developers lack platforms provided readily and at no price to validate their solution to determine the best hardware for their specific needs.
The Reference Implementation
For demonstration purposes, Intel trained a BTS U-Net model using BRATS dataset.
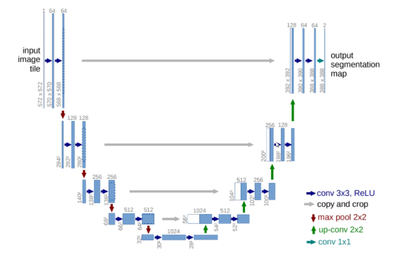
U-Net is a convolutional DNN developed for biomedical image segmentation; it can segment images using a limited amount of training data. The U-Net architecture has known an immense success in the biomedical community, making it the primary tool for segmentation. Its usage is widespread across 2D and 3D biomedical modalities from ultrasound, and X-ray images to MRI and CT images. To this day, the original U-Net paper by O. Ronneberger [1] has been cited more than 40000 times. The U-Net success is due to the modularity of the architecture, where numerous variations can be brought to the low-level architecture while keeping the high-level U-shape.
Figure 1: U-Net Architecture Diagram
Therefore, we are showcasing in this article more than just a use-case on a precise modality. We are demonstrating how a gold-standard architecture, widely used in the medical imaging domain, can be optimized for Intel architecture, regardless of the modality it is applied to. In this example, the U-Net model was trained using Keras/TensorFlow.
To train the U-Net, the dataset BRATS is used. BRATS stands for ‘The Multimodal Brain Tumor Image Segmentation Benchmark’ and it is a set of 65 multi-contrast MR scans of low and high-grade glioma patients, manually annotated by up to four raters and thus provides an established ground-truth tumor segmentation.
After training, we converted the Keras/TensorFlow model using OpenVINO Model Optimizer, this provides a first stage of hardware-agnostic graph optimizations, that makes the model leaner. Then, when running the inference using Inference Engine API, the built-in hardware plugin on which the model is being run on (CPU plugin, GPU plugin, …) will also come with a set of runtime optimizations that provides additional speed-ups. Therefore, when comparing the CPU inference performance on Keras/Tensorflow vs OpenVINO, OpenVINO showcases performance improvement. For other hardware, OpenVINO provides better performance/watts. The switch of hardware plugins is very easy in OpenVINO, as it is only a flag to set in the API.
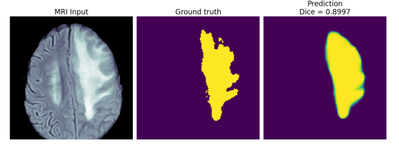
OpenVINO optimization also makes models interoperable, and inference could be in run on numerous Intel Architecture as the OpenVINO Inference Engine uses built-in plug-ins, thereby removing the need for extensive architecture-based coding. Interoperability also allows heterogenous computing like running differ layers of the U-Net topology on different hardware using HETERO command to decrease latency or running the model on multiple hardware using MULTI command to increase throughput. Interoperability gives developers the ability to choose the right hardware but practically it would be tough for each developer to have a large lab with this range of hardware to test. Hence, we have provided the BTS model on the Intel DevCloud for Edge as a sample application. DevCloud for edge provides a large cluster of hardware free of cost and the sample application provides code example on how to optimize the model, run the model on different hardware (edge nodes). The sample application compares the performance of the OpenVINO optimized model with the original Keras/Tensorflow model and the segmentation results with ground truth to calculate Sørensen–Dice coefficient. See Figure 1 for an example segmentation of an MRI slice by OpenVINO.
Figure 2 Segmentation of MRI slice in the DevCloud for the Edge BTS sample (right is the prediction after inference by OpenVINO)
This application can act as a reference application or base code for developers to implement OpenVINO optimized features and run their workloads on DevCloud architecture.
Intel went further and reused the trained model and BTS sample application on DevCloud for edge to build a BTS Reference Implementation (RI) on the Intel Developer Catalog. BTS RI is implemented as a container-based solution running on Kubernetes. It uses Grafana dashboard to display the performance comparison between Keras/TensorFlow and OpenVINO optimized model as well as hardware telemetry. Developer can view the power of OpenVINO by just running an installation file which will implement the BTS solution even on their Intel Core based laptop and they will be able to view efficient U-Net based segmentation on MR images.
Conclusion
OpenVINO model optimization, and platforms like DevCloud for Edge and Intel Developer Catalog provides tools in the developer’s hand to deploy Medical Imaging AI workloads such as ‘Brain Tumor Segmentation using U-Net topology’ easily, efficiently and at scale on Intel Architecture.
About OpenVINO™
The Intel® Distribution of OpenVINO™ is an open-source toolkit for high-performance, deep learning inferencing for ultra-fast, amazingly accurate real-world results deployed across Intel® hardware from edge to cloud. Therefore, simplifying the path to inferencing with a streamlined development workflow. Developers can write an application or algorithm once and deploy it across Intel® architecture, including CPU, GPU, iGPU, Movidius VPU, and GNA.
Citations:
[1] Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
Resources:
• Get started with OpenVINO™: https://docs.openvinotoolkit.org/latest/index.html
- Intel's AI and Deep Learning Solutions: https://www.intel.com/content/www/us/en/artificial-intelligence/overview.html
You must be a registered user to add a comment. If you've already registered, sign in. Otherwise, register and sign in.